The role of synovial fibroblasts in RA
Synovial fibroblasts are the major stromal cells in the joint synovium and play a crucial part in inflammation and joint destruction in RA.
Synovial fibroblasts from patients with RA exhibit an intrinsically activated phenotype, which is maintained under cell culture conditions and is characterised by increased expression of matrix-degrading enzymes and pro-inflammatory cytokines and chemokines. This aggressive phenotype is believed to be sustained by alterations in epigenetic modifications, such as DNA methylation and non-coding RNA. In our work, we look at different aspects of fibroblast biology to decipher how these cells are activated in RA, how they contribute to disease and how they can be therapeutically targeted.
Current projects
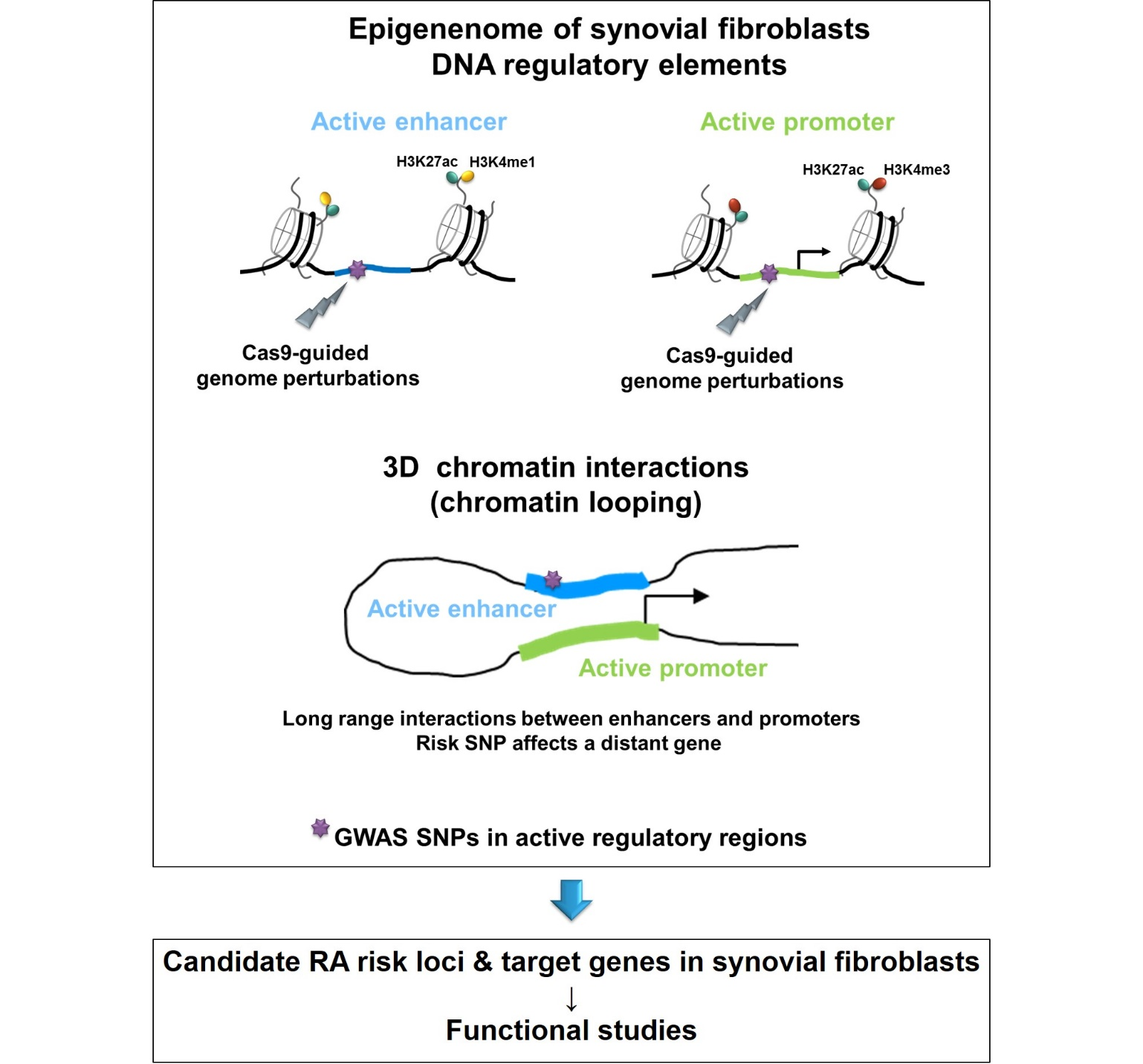
Genetic factors contribute 50% to the risk of RA and the heritability of RA is estimated to be approximately 60%. Genome-wide association studies (GWAS) have identified over 100 genetic susceptibility loci for RA. However, only a small proportion of RA risk loci have been functionally analysed to date. In this project, we align genetic data with in depth analysis of the epigenetic landscape of synovial fibroblasts to determine which genetic risk loci lie within active, regulatory DNA regions in synovial fibroblasts. We then further analyse these DNA regions in synovial fibroblasts to see the molecular and functional impact of the genetic risk loci on fibroblast biology.
Involved team members
- Miranda Houtman
- Larissa Moser
Funded by
- Swiss National Science Foundation
Collaborators
Prof Stephen Eyre, Arthritis Research UK Centre for Genetics and Genomics, University of Manchester, UK
Relevant Publications
- Ge X, Frank-Bertoncelj M, Klein K, McGovern A, Kuret T, Houtman M, Burja B, Micheroli R, Shi C, Marks M, Filer A, Buckley CD, Orozco G, Distler O, Morris AP, Martin P, Eyre S, Ospelt C. Functional genomics atlas of synovial fibroblasts defining rheumatoid arthritis heritability. Genome Biol. 2021 Aug 25;22(1):247. doi: 10.1186/s13059-021-02460-6.
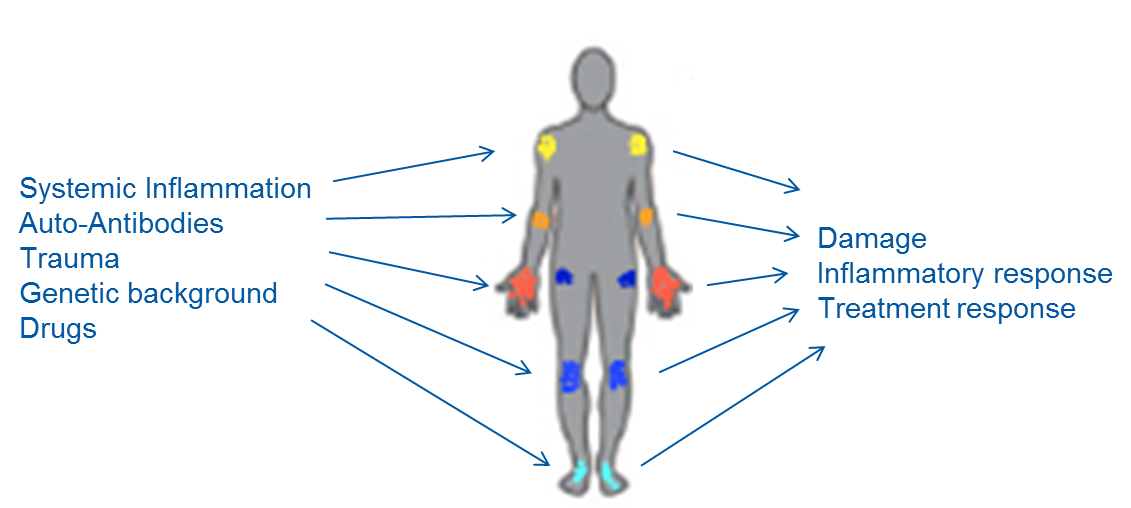
We could show that synovial fibroblasts significantly differ between different joint locations in their epigenome, transcriptome and function. We hypothesize that the differences in these local cells of the joints substantially influence the inflammatory response, the influx of immune cells, the resolution of inflammation and thus disease outcome and therapeutic response. To test this hypothesis, we analyze joint-specific differences in the inflammatory and anti-inflammatory response using histological and single cell molecular analysis and try to identify the key regulators of joint-specific inflammatory signaling pathways.
Involved team members
- Eva Camarillo
- Muriel Elhai
- Miranda Houtman
- Raphael Micheroli
Funded by
- Iten-Kohaut-Foundation
- Novartis Foundation for medical-biological research
- MLR Foundation
Relevant publications
- Frank-Bertoncelj M, Trenkmann M, Klein K, Karouzakis E, Rehrauer H, Bratus A, Kolling C, Armaka M, Filer A, Michel BA, Gay RE, Buckley CD, Kollias G, Gay S, Ospelt C. Epigenetically-driven anatomical diversity of synovial fibroblasts guides joint-specific fibroblast functions. Nat. Commun. 8, 14852 doi: 10.1038/ncomms14852 (2017)
- Ospelt C, Frank-Bertoncelj M. Why location matters – site-specific factors in rheumatic diseases. Nat Rev Rheumatol. 2017 Jul;13(7):433-442. doi: 10.1038/nrrheum.2017.96.
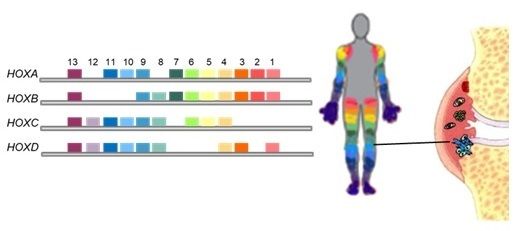
We have shown that synovial fibroblasts isolated from different joints substantially differ in gene expression and functional properties. In particular, the expression of genes that are involved in embryonic limb development, namely HOX genes, substantially differs between joint regions. Also in skin and cartilage, the expression of HOX transcription factors is strongly related to the anatomical localisation. Based on this data, we hypothesize that the joint-specific phenotypes shaped during embryonic development and maintained during adult life, significantly contribute to the pathognomic patterns of joint affection in chronic inflammatory arthritis such as RA. However, little is known about the function of HOX genes in synovial fibroblasts. Therefore, we analyse the properties of various HOX transcription factors and non-coding RNAs in synovial fibroblasts with knock-down and overexpression experiments.
Involved team members
- Eva Camarillo
- Muriel Elhai
- Masoumeh Mirrahimi
- Larissa Moser
Funded by
- Foundation for Research in Science and the Humanities at the University of Zurich
- EMDO Foundation
- Hartmann-Müller Foundation
- Kurt-und-Senta-Herrmann Foundation
- Carigest
Relevant publications
- Elhai M, Micheroli R, Frank-Bertoncelj M, Klein K, Distler O, Ospelt C.
The Long Non-coding RNA HOTAIR Regulates BMP2 and Wnt Pathways in Synovial Fibroblasts [abstract].
Arthritis Rheumatol. 2019; 71 (suppl 10). - Frank-Bertoncelj M, Kuret T, Županič A, Sodin-Semrl S, Distler O, Ospelt C.
The Long Noncoding RNA HOTTIP Regulates Cell Cycle and Inflammatory Response in Hand Synovial Fibroblasts [abstract].
Arthritis Rheumatol. 2019; 71 (suppl 10).








