Cellular Therapy and Leukemia Research – Schneidawind Lab
Keywords
Disease models, acute leukemia, genome engineering, cell therapy, immune tolerance
Summary & Mission statement
We focus on understanding the biology of leukemogenesis and the immunobiology of hematopoietic cell transplantation. Our goal is to establish patient-specific disease models to find novel therapeutic approachs to cure leukemia and to improve efficacy and safety of cell therapies.
Overview
Our lab is interested in novel gene editing techniques like CRISPR/Cas9 to understand the pathobiology of hematological diseases. Therefore, our lab developed human BCR/ABL and MLL rearranged models based on patient-specific translocations induced in hematopoietic stem and progenitor cells (HSPCs) derived from both cord blood and bone marrow. By using different microenvironments, we promote lymphatic and myeloid differentiation. These genome-edited cells authentically mimic adult and childhood patient disease. By using genetic analyses, we gain valuable knowledge about disease mechanisms uncovering novel therapeutic targets. Candidate compounds can be easily uncovered and tested with our platforms allowing us to understand their mode of action.
Another interest of our lab is the optimization of cell therapies using chimeric antigen receptor technologies targeting novel antigens for the treatment of acute leukemia. Hereby, we focus on particular effector cell populations such as invariant natural killer T (iNKT) cells. iNKT cells are potent regulators of immune responses with strong cytotoxic properties that can be further increased by introducing chimeric antigen receptors. iNKT cells also prevent graft-versus-host disease after allogeneic hematopoietic cell transplantation (HCT). We develop novel cytotherapeutic approaches to improve outcomes after allogeneic HCT by reducing relapse rates and non-relapse mortality.
Publications
Figure
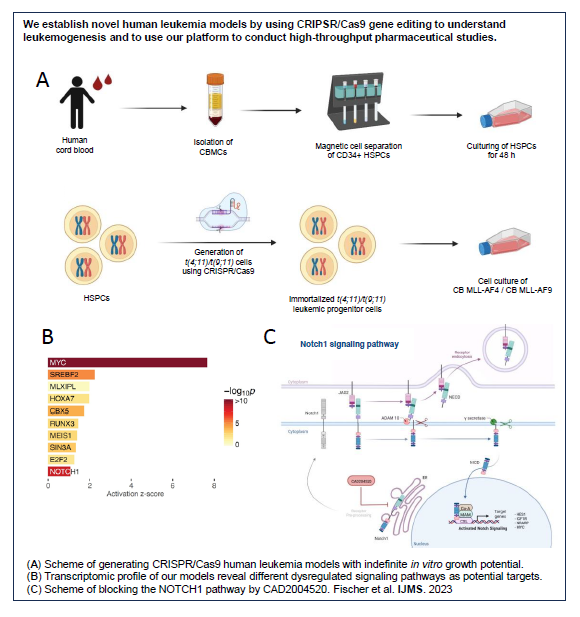
A) Scheme of generating CRISPR/Cas9 human leukemia models with indefinite in vitro growth potential. (B) Transcriptomic profile of our models reveal different dysregulated signaling pathways as potential targets. (C) Scheme of blocking the NOTCH1 pathway by CAD2004520. Fischer et al. International Journal of Molecular Sciences 2023
