Research Group Hanspeter Naegeli
Our research is dedicated to the study of signaling, protein modification and chromatin remodeling pathways that promote genome stability.
Keywords
DNA repair, histone modifiers, chromatin remodelers
Summary & Mission statement
Our research is dedicated to the study of signaling, protein modification and chromatin remodeling pathways that promote genome stability. We are currently focusing on the mechanisms by which histone methyltransferases and chromatin remodelers regulate nucleotide excision repair activity in human cells.
Overview
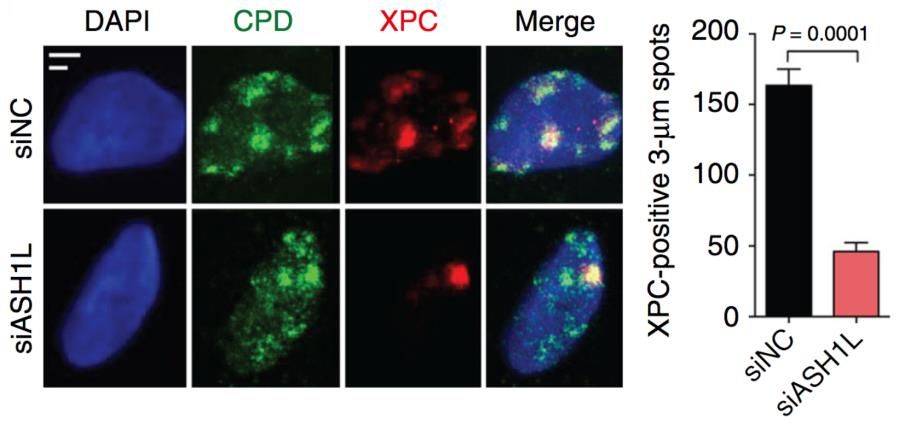
Our aim is to understand how the histone methylome allows for DNA damage recognition by the nucleotide excision repair (NER) “big enzyme” machine in human cells. Maintenance of genome stability is essential to prevent cancer and premature aging. The NER machine safeguards the DNA double helix by removing bulky base adducts induced by ultraviolet (UV) radiation, chemical carcinogens, metabolic byproducts and oxygen radicals. The most prevalent bulky lesion arises from the UV spectrum of sunlight or artificial devices, which can induce hundreds of thousands of DNA photoproducts in each skin cell. To be effective, the NER machine must cope with the inherent compaction of its chromatin substrate where DNA is wrapped around histones giving rise to nucleosome arrays. Work in our laboratory demonstrated a novel role of the histone methyltransferase ASH1L in guiding the NER machine to packed chromatin. We are now using protein depletions and gene deletions as well as chromatin immunoprecipitations followed by proteomics analyses to determine the role of (i) methyltransferases that add chromatin-relaxing methyl groups to histones (for example to H3Lys4), (ii) methyltransferases that add chromatin-condensing methyl groups to histones (for example to H3Lys9) and (iii) chromatin remodelers recruited or activated by histone methylation.
Publications